Please use this identifier to cite or link to this item:
https://doi.org/10.21256/zhaw-23864| Publication type: | Article in scientific journal |
| Type of review: | Peer review (publication) |
| Title: | Xanthomonas hydrangeae sp. nov., a novel plant pathogen isolated from Hydrangea arborescens |
| Authors: | Dia, Nay C. Van Vaerenbergh, Johan Van Malderghem, Cinzia Blom, Jochen Smits, Theo H. M. Cottyn, Bart Pothier, Joël F. |
| et. al: | No |
| DOI: | 10.1099/ijsem.0.005163 10.21256/zhaw-23864 |
| Published in: | International Journal of Systematic and Evolutionary Microbiology |
| Volume(Issue): | 71 |
| Issue: | 12 |
| Issue Date: | 16-Dec-2021 |
| Publisher / Ed. Institution: | Microbiology Society |
| ISSN: | 1466-5026 1466-5034 |
| Language: | English |
| Subjects: | Xanthomonas; diagnostics; Overall genome-relatedness indices |
| Subject (DDC): | 572: Biochemistry 579: Microbiology |
| Abstract: | This paper describes a novel species isolated in 2011 and 2012 from nursery-grown Hydrangea arborescens cultivars in Flanders, Belgium. After 4 days at 28 °C, the strains yielded yellow, round, convex and mucoid colonies. Pathogenicity of the strains was confirmed on its isolation host, as well as on Hydrangea quercifolia. Analysis using MALDI-TOF MS identified the Hydrangea strains as belonging to the genus Xanthomonas but excluded them from the species Xanthomonas hortorum. A phylogenetic tree based on gyrB confirmed the close relation to X. hortorum. Three fatty acids were dominant in the Hydrangea isolates: anteiso-C15 : 0, iso-C15 : 0 and summed feature 3 (C16 : 1 ω7c/C16 : 1 ω6c). Unlike X. hortorum pathovars, the Hydrangea strains were unable to grow in the presence of lithium chloride and could only weakly utilize d-fructose-6-PO4 and glucuronamide. Phylogenetic characterization based on multilocus sequence analysis and phylogenomic characterization revealed that the strains are close to, yet distinct from, X. hortorum. The genome sequences of the strains had average nucleotide identity values ranging from 94.35-95.19 % and in silico DNA-DNA hybridization values ranging from 55.70 to 59.40 % to genomes of the X. hortorum pathovars. A genomics-based loop-mediated isothermal amplification assay was developed which was specific to the Hydrangea strains for its early detection. A novel species, Xanthomonas hydrangeae sp. nov., is proposed with strain LMG 31884T (=CCOS 1956T) as the type strain. |
| URI: | https://digitalcollection.zhaw.ch/handle/11475/23864 |
| Related research data: | https://www.ebi.ac.uk/ena/browser/view/PRJEB38813 |
| Fulltext version: | Accepted version |
| License (according to publishing contract): | CC BY 1.0: Attribution 1.0 Generic |
| Departement: | Life Sciences and Facility Management |
| Organisational Unit: | Institute of Natural Resource Sciences (IUNR) |
| Published as part of the ZHAW project: | Xhortomics: Entwicklung von diagnostischen und epidemiologischen Hilfsmitteln für die Xanthomonas hortorum Spezies Gruppe basierend auf OMICs Technologien |
| Appears in collections: | Publikationen Life Sciences und Facility Management |
Files in This Item:
| File | Description | Size | Format | |
|---|---|---|---|---|
| Dia-etal_Xanthomonas-hydrangeae_IJSEM-Accepted-Version.pdf | Accepted Version | 306 kB | Adobe PDF |  View/Open |
| Dia-etal_Xanthomonas-hydrangeae_IJSEM-Supplementary-Material-Accepted-Version.pdf | Accepted Supplementary Material | 582.24 kB | Adobe PDF |  View/Open |
| Fig1-Disease-symptoms.png | 4.27 MB | image/png | 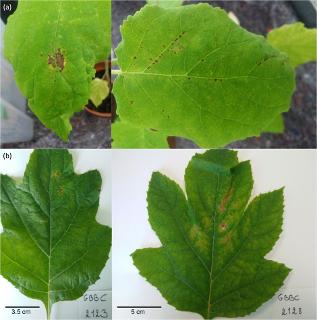 View/Open | |
| Fig2-16S-tree.eps | 4.45 MB | Postscript | View/Open | |
| Fig3-MLSA-first-trim-tree.eps | 2.12 MB | Postscript | View/Open | |
| Fig4-whole-genome-tree.eps | 2.02 MB | Postscript | View/Open | |
| Fig5-ANI-isDDH.png | 1.33 MB | image/png |  View/Open |
Show full item record
Dia, N. C., Van Vaerenbergh, J., Van Malderghem, C., Blom, J., Smits, T. H. M., Cottyn, B., & Pothier, J. F. (2021). Xanthomonas hydrangeae sp. nov., a novel plant pathogen isolated from Hydrangea arborescens. International Journal of Systematic and Evolutionary Microbiology, 71(12). https://doi.org/10.1099/ijsem.0.005163
Dia, N.C. et al. (2021) ‘Xanthomonas hydrangeae sp. nov., a novel plant pathogen isolated from Hydrangea arborescens’, International Journal of Systematic and Evolutionary Microbiology, 71(12). Available at: https://doi.org/10.1099/ijsem.0.005163.
N. C. Dia et al., “Xanthomonas hydrangeae sp. nov., a novel plant pathogen isolated from Hydrangea arborescens,” International Journal of Systematic and Evolutionary Microbiology, vol. 71, no. 12, Dec. 2021, doi: 10.1099/ijsem.0.005163.
DIA, Nay C., Johan VAN VAERENBERGH, Cinzia VAN MALDERGHEM, Jochen BLOM, Theo H. M. SMITS, Bart COTTYN und Joël F. POTHIER, 2021. Xanthomonas hydrangeae sp. nov., a novel plant pathogen isolated from Hydrangea arborescens. International Journal of Systematic and Evolutionary Microbiology. 16 Dezember 2021. Bd. 71, Nr. 12. DOI 10.1099/ijsem.0.005163
Dia, Nay C., Johan Van Vaerenbergh, Cinzia Van Malderghem, Jochen Blom, Theo H. M. Smits, Bart Cottyn, and Joël F. Pothier. 2021. “Xanthomonas Hydrangeae Sp. Nov., A Novel Plant Pathogen Isolated from Hydrangea Arborescens.” International Journal of Systematic and Evolutionary Microbiology 71 (12). https://doi.org/10.1099/ijsem.0.005163.
Dia, Nay C., et al. “Xanthomonas Hydrangeae Sp. Nov., A Novel Plant Pathogen Isolated from Hydrangea Arborescens.” International Journal of Systematic and Evolutionary Microbiology, vol. 71, no. 12, Dec. 2021, https://doi.org/10.1099/ijsem.0.005163.
Items in DSpace are protected by copyright, with all rights reserved, unless otherwise indicated.